Ecologia Numérica
Aula 2 - Descrição de comunidades ecológicas
Felipe Melo
Laboratório de Ecologia Aplicada - UFPE
2022-12-11
1 / 17
Estrutura de dados ecológicos
- Bases de dados ecológicos seguem uma lógica relativamente simples
- As bases são organizadas quse sempre na forma de matrizes com 'n' colunas e 'p' linhas
## # A tibble: 10 × 5## spec siteA siteB siteC siteD## <chr> <int> <int> <int> <int>## 1 sp1 0 0 1 0## 2 sp2 1 0 0 0## 3 sp3 0 1 0 1## 4 sp4 0 0 0 1## 5 sp5 0 0 1 1## 6 sp6 1 0 1 1## 7 sp7 0 1 0 0## 8 sp8 1 1 0 0## 9 sp9 1 0 0 0## 10 sp10 1 0 0 0Todas as bases ecológicas que iremos usar serão organizadas dessa forma
2 / 17
Peixes de um rio na Suíça
- Usamos a mesma lógica para organizar bas bases de dados de espécies
- Por exemplo, essa é a estrutura dos dados ambientais que usaremos nesse módulo
Estão no livro Numerical Ecology with R
- Por exemplo, essa é a estrutura dos dados ambientais que usaremos nesse módulo
## Cogo Satr Phph Babl Thth Teso Chna Pato Lele Sqce Baba Albi Gogo Eslu Pefl Rham Legi Scer Cyca Titi## 1 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0## 2 0 5 4 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0## 3 0 5 5 5 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0## 4 0 4 5 5 0 0 0 0 0 1 0 0 1 2 2 0 0 0 0 1## 5 0 2 3 2 0 0 0 0 5 2 0 0 2 4 4 0 0 2 0 3## 6 0 3 4 5 0 0 0 0 1 2 0 0 1 1 1 0 0 0 0 2## 7 0 5 4 5 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0## 8 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0## 9 0 0 1 3 0 0 0 0 0 5 0 0 0 0 0 0 0 0 0 1## 10 0 1 4 4 0 0 0 0 2 2 0 0 1 0 0 0 0 0 0 0## 11 1 3 4 1 1 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0## 12 2 5 4 4 2 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0## 13 2 5 5 2 3 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0## 14 3 5 5 4 4 3 0 0 0 1 1 0 1 1 0 0 0 0 0 0## 15 3 4 4 5 2 4 0 0 3 3 2 0 2 0 0 0 0 0 0 1## 16 2 3 3 5 0 5 0 4 5 2 2 1 2 1 1 0 1 0 1 1## 17 1 2 4 4 1 2 1 4 3 2 3 4 1 1 2 1 1 0 1 1## 18 1 1 3 3 1 1 1 3 2 3 3 3 2 1 3 2 1 0 1 1## 19 0 0 3 5 0 1 2 3 2 1 2 2 4 1 1 2 1 1 1 2## 20 0 0 1 2 0 0 2 2 2 3 4 3 4 2 2 3 2 2 1 4## 21 0 0 1 1 0 0 2 2 2 2 4 2 5 3 3 3 2 2 2 4## 22 0 0 0 1 0 0 3 2 3 4 5 1 5 3 4 3 3 2 3 4## 23 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0## 24 0 0 0 0 0 0 1 0 0 2 0 0 1 0 0 0 1 0 0 0## 25 0 0 0 0 0 0 0 0 1 1 0 0 2 1 0 0 0 1 0 0## 26 0 0 0 1 0 0 1 0 1 2 2 1 3 2 1 2 2 1 1 3## 27 0 0 0 1 0 0 1 1 2 3 4 1 4 4 1 3 3 1 2 5## 28 0 0 0 1 0 0 1 1 2 4 3 1 4 3 2 4 4 2 4 4## 29 0 1 1 1 1 1 2 2 3 4 5 3 5 5 4 5 5 2 3 3## 30 0 0 0 0 0 0 1 2 3 3 3 5 5 4 5 5 3 5 5 5## Abbr Icme Gyce Ruru Blbj Alal Anan## 1 0 0 0 0 0 0 0## 2 0 0 0 0 0 0 0## 3 0 0 0 0 0 0 0## 4 0 0 0 0 0 0 0## 5 0 0 0 5 0 0 0## 6 0 0 0 1 0 0 0## 7 0 0 0 0 0 0 0## 8 0 0 0 0 0 0 0## 9 0 0 0 4 0 0 0## 10 0 0 0 0 0 0 0## 11 0 0 0 0 0 0 0## 12 0 0 0 0 0 0 0## 13 0 0 0 0 0 0 0## 14 0 0 0 0 0 0 0## 15 0 0 0 0 0 0 0## 16 0 0 0 1 0 0 0## 17 0 0 0 2 0 2 1## 18 0 0 1 2 0 2 1## 19 1 0 1 5 1 3 1## 20 1 0 2 5 2 5 2## 21 3 1 3 5 3 5 2## 22 4 2 4 5 4 5 2## 23 0 0 0 1 0 2 0## 24 0 0 2 2 1 5 0## 25 0 0 1 1 0 3 0## 26 2 1 4 4 2 5 2## 27 3 2 5 5 4 5 3## 28 3 3 5 5 5 5 4## 29 4 4 5 5 4 5 4## 30 5 5 5 5 5 5 53 / 17
Matriz ambietal
- Usamos a mesma lógica para organizar bas bases de dados ambientais
- Por exemplo, essa é a estrutura dos dados ambientais que usaremos nesse módulo
Estão no livro Numerical Ecology with R
- Por exemplo, essa é a estrutura dos dados ambientais que usaremos nesse módulo
## dfs ele slo dis pH har pho nit amm oxy bod## 1 0.3 934 48.0 0.84 7.9 45 0.01 0.20 0.00 12.2 2.7## 2 2.2 932 3.0 1.00 8.0 40 0.02 0.20 0.10 10.3 1.9## 3 10.2 914 3.7 1.80 8.3 52 0.05 0.22 0.05 10.5 3.5## 4 18.5 854 3.2 2.53 8.0 72 0.10 0.21 0.00 11.0 1.3## 5 21.5 849 2.3 2.64 8.1 84 0.38 0.52 0.20 8.0 6.2## 6 32.4 846 3.2 2.86 7.9 60 0.20 0.15 0.00 10.2 5.34 / 17
Matriz espacial
- Usamos a mesma lógica para organizar bas bases de dados espaciais
- Por exemplo, essa é a estrutura dos dados ambientais que usaremos nesse módulo
Estão no livro Numerical Ecology with R
- Por exemplo, essa é a estrutura dos dados ambientais que usaremos nesse módulo
## X Y## 1 85.678 20.000## 2 84.955 20.100## 3 92.301 23.796## 4 91.280 26.431## 5 92.005 29.163## 6 95.954 36.3155 / 17
Vamos olhar mais de perto para a base de dados de espécies
nrow(spe) # entendendo a base, sabemos que o número de linhas equivale ao de amostras## [1] 30ncol(spe) # o número de colunas equivale ao de espécies## [1] 27dim(spe) # esse comando já dá tudo## [1] 30 27colnames(spe) # aqui até os nomes abravidaos das espécies nós temos## [1] "Cogo" "Satr" "Phph" "Babl" "Thth" "Teso" "Chna" "Pato" "Lele" "Sqce" "Baba" "Albi" "Gogo" "Eslu"## [15] "Pefl" "Rham" "Legi" "Scer" "Cyca" "Titi" "Abbr" "Icme" "Gyce" "Ruru" "Blbj" "Alal" "Anan"6 / 17
Essa regra vale para todas as bases
Pratique com todas as bases
- Cheque a estrutura
- Confira a natureza das variáveis
- Não avance antes de ter certeza que você conhece seus dados
- Os dados são amigos, NÃO OS TORTURE
7 / 17
Faça alguns gráficos
- Gráficos precisam estar acompanhados de pergumtas
- Qual a distribuição de abundância das espécies?
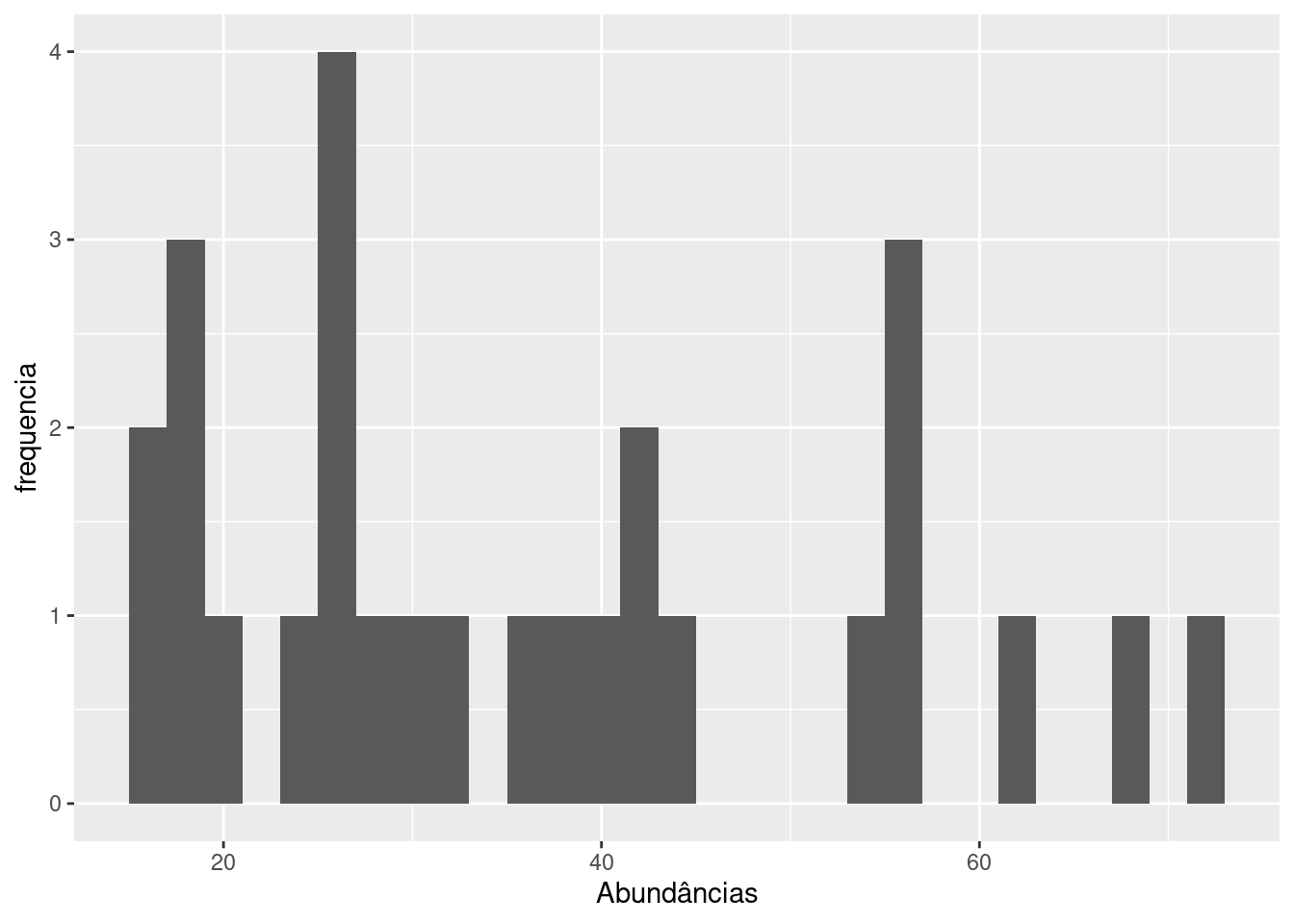
- Este é um gráfico muito útil, que auxilia muito na compreensão da diversidade biológica de comunidades
8 / 17
Faça mais gráficos
- Entenda a amplitude das variáveis ambientais que possui
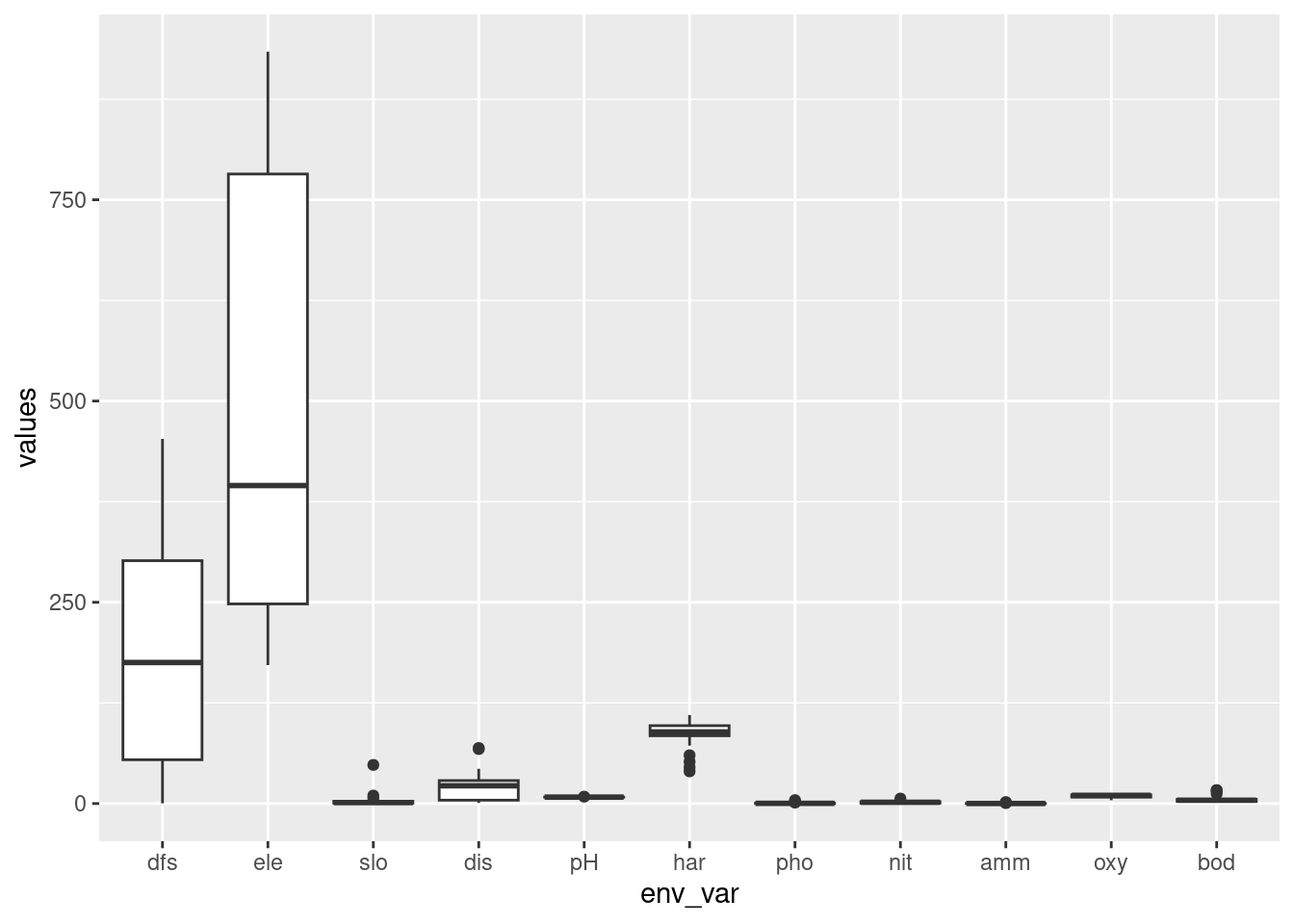
- Note que as variáveis têm "amplitudes" diferentes, e isso é importante
9 / 17
Entenda também suas variáveis espaciais
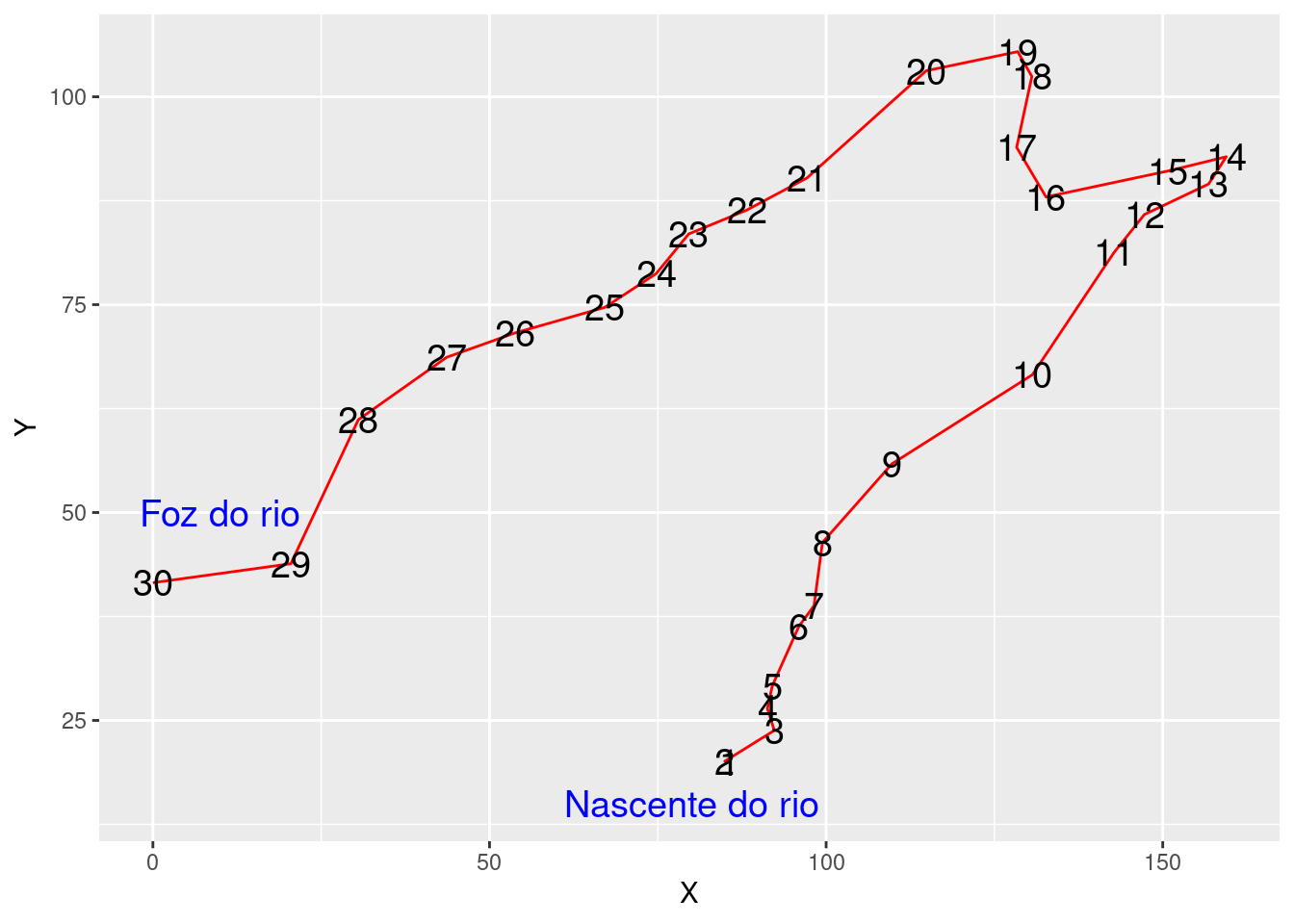
Essa é uma representação do rio onde as espécies foram coletadas
10 / 17
Misture as matrizes
Espaço + Ambiente
Espaço + Ambiente + Dados biológicos
11 / 17
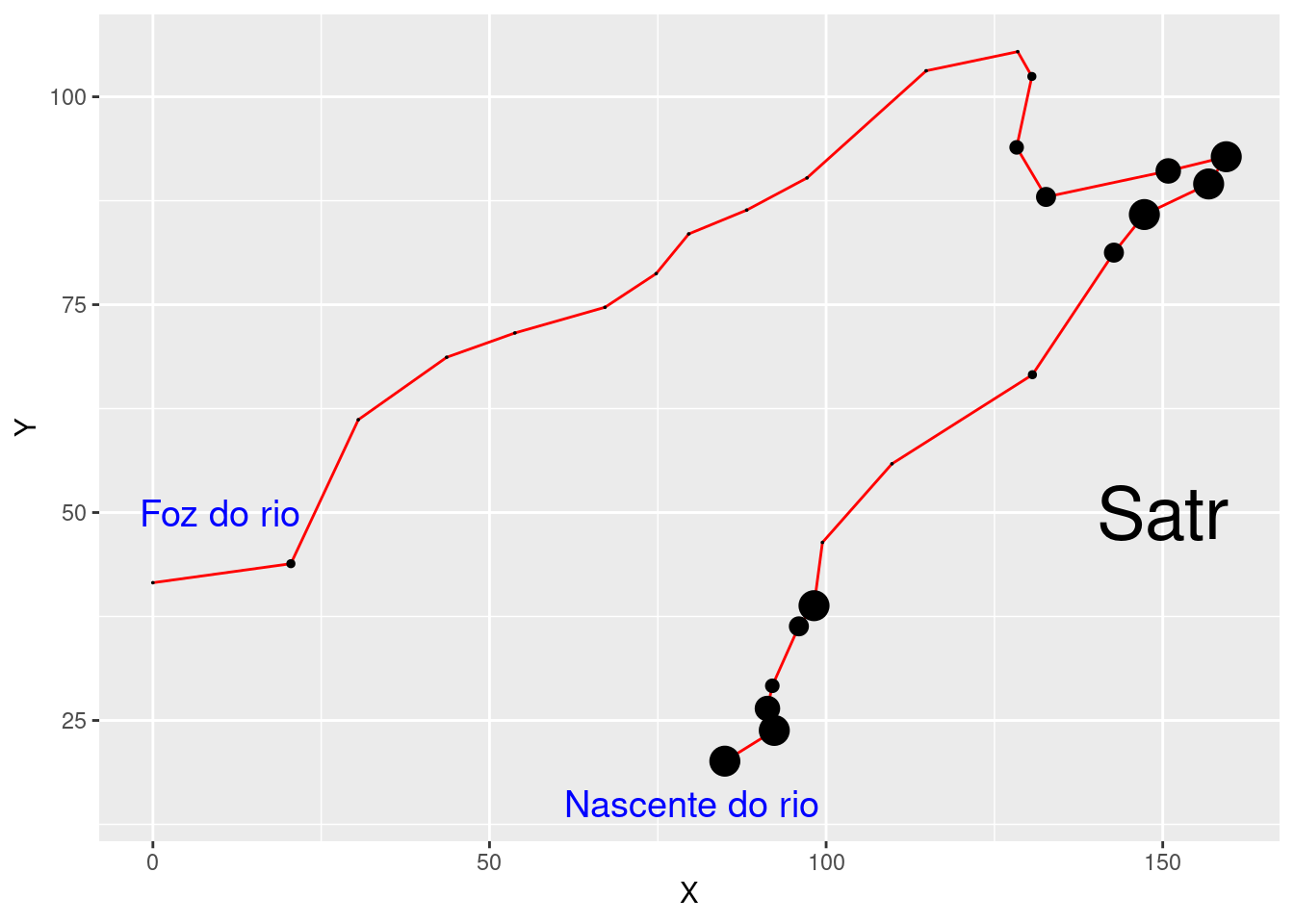
O processo de namoro dos dados nunca termina...
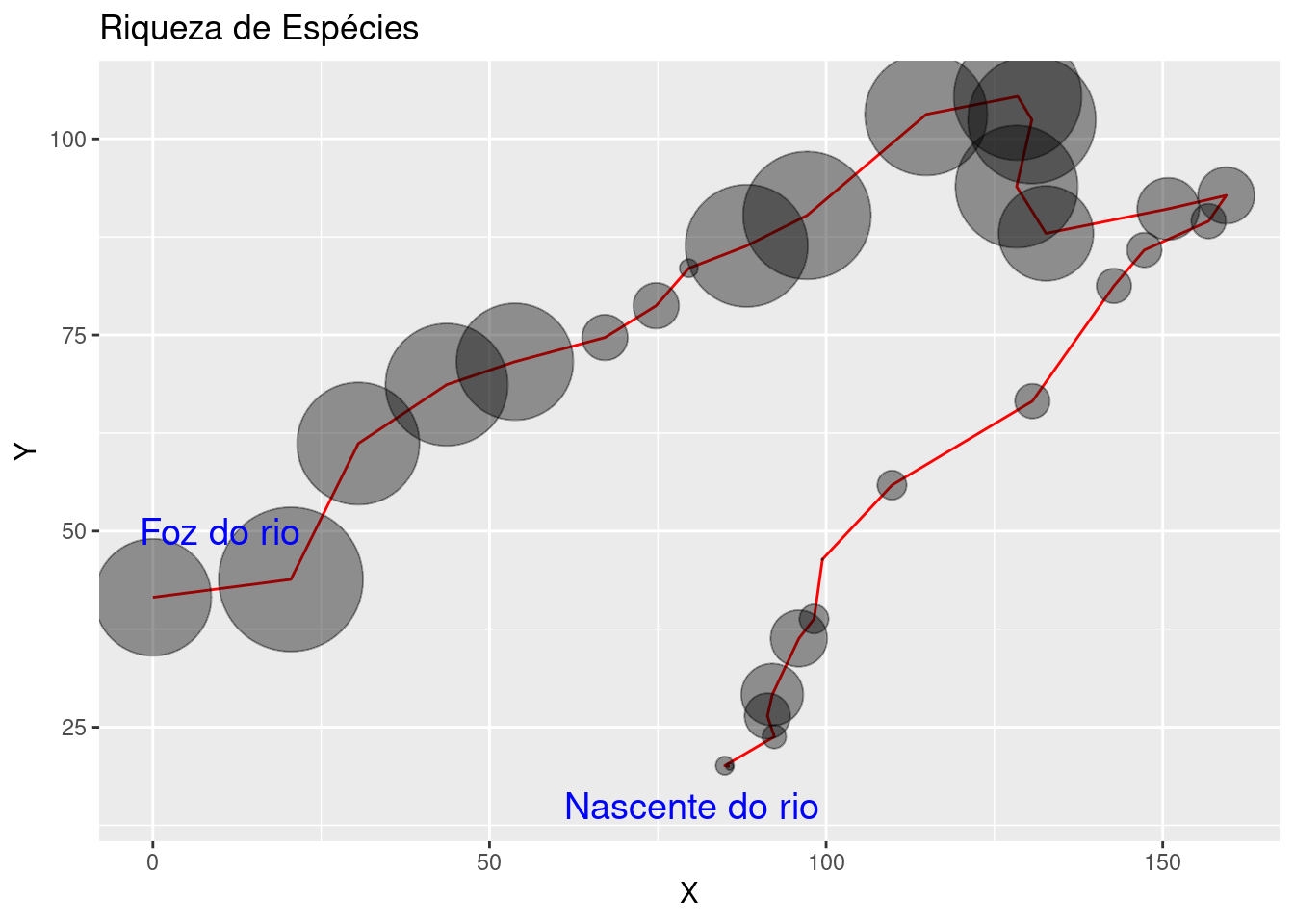
- note os valores de riqueza representados pelo tamanho dos pontos
12 / 17
O processo de namoro dos dados nunca termina...

- note os valores de riqueza representados pelo tamanho dos pontos
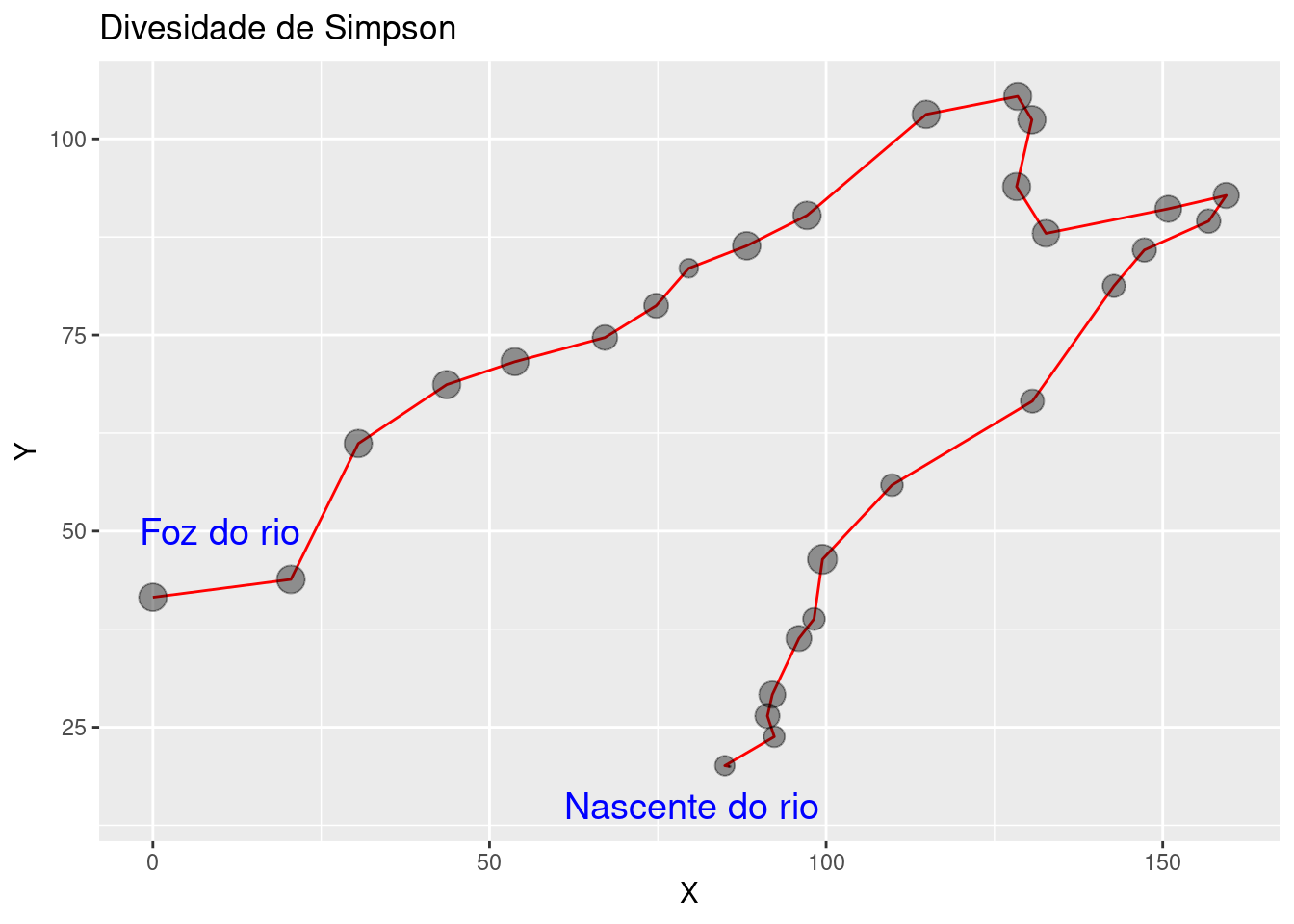
- Simpson parace menos sensível
12 / 17
Como é a distribuição de abundância dessa comunidade de peixes?
Código
## Cogo Satr Phph Babl Thth Teso Chna Pato Lele Sqce## 1 0 3 0 0 0 0 0 0 0 0## 2 0 5 4 3 0 0 0 0 0 0## 3 0 5 5 5 0 0 0 0 0 0## 4 0 4 5 5 0 0 0 0 0 1## 5 0 2 3 2 0 0 0 0 5 2## 6 0 3 4 5 0 0 0 0 1 2## 7 0 5 4 5 0 0 0 0 1 1## 8 0 0 0 0 0 0 0 0 0 0## 9 0 0 1 3 0 0 0 0 0 5## 10 0 1 4 4 0 0 0 0 2 214 / 17
Plot

15 / 17
Plot melhorado
## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.## ℹ Please use `linewidth` instead.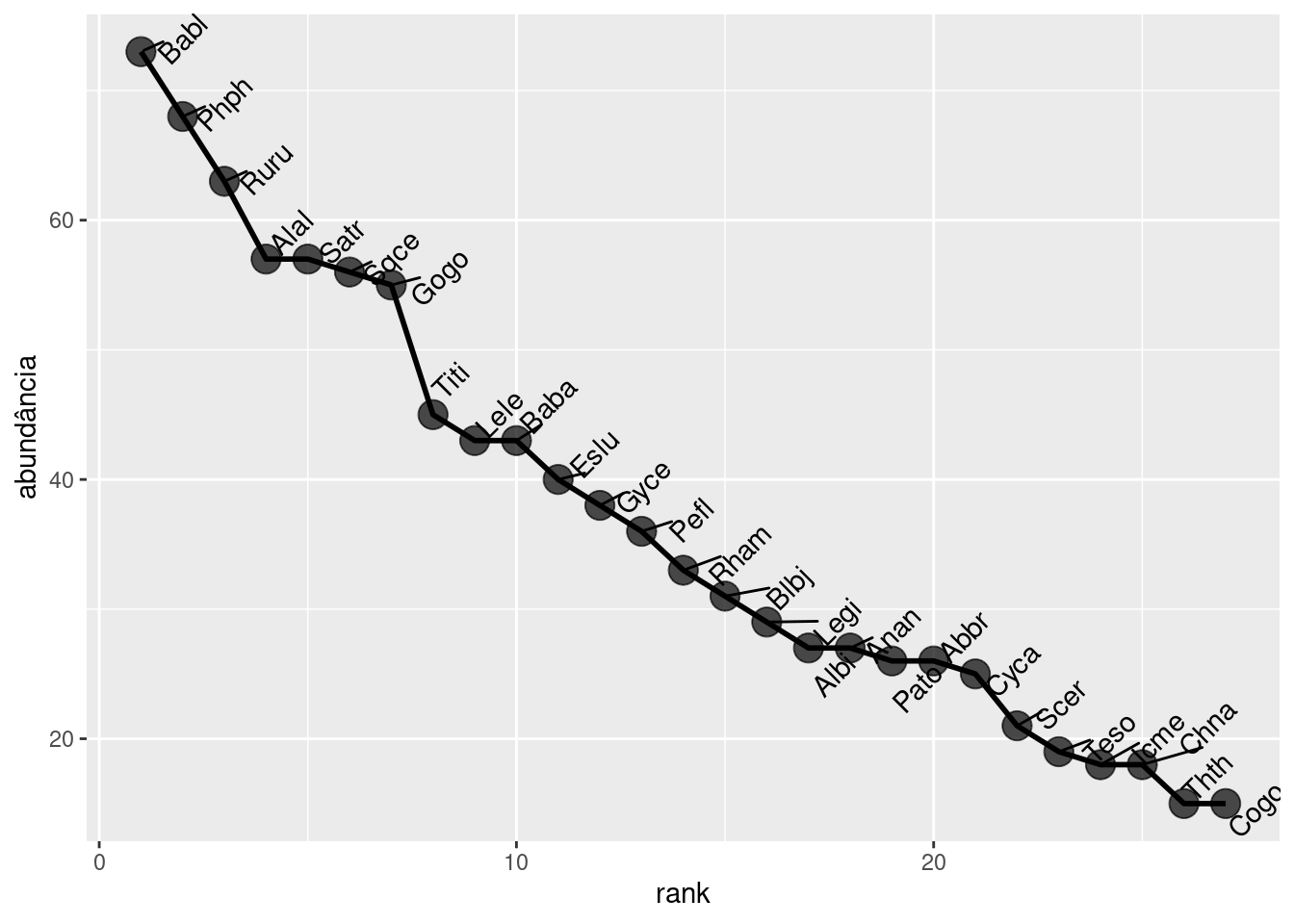
16 / 17
Faça exercícios, sempre utilizando perguntas originais
- Use os dados fornecidos pelo livro Numerical Ecology with R
- Mas... Evite apenas seguir os scripts e exemplos
- Monte seu site usando o RMarkdown
- Não desista, isso é treino

17 / 17
